Allen V1 Model: Large-scale Network
Overview
This tutorial demonstrates a practical example of building a mouse primary visual cortex column model with bionetlite. This is a large-scale biophysical model used in actual research.
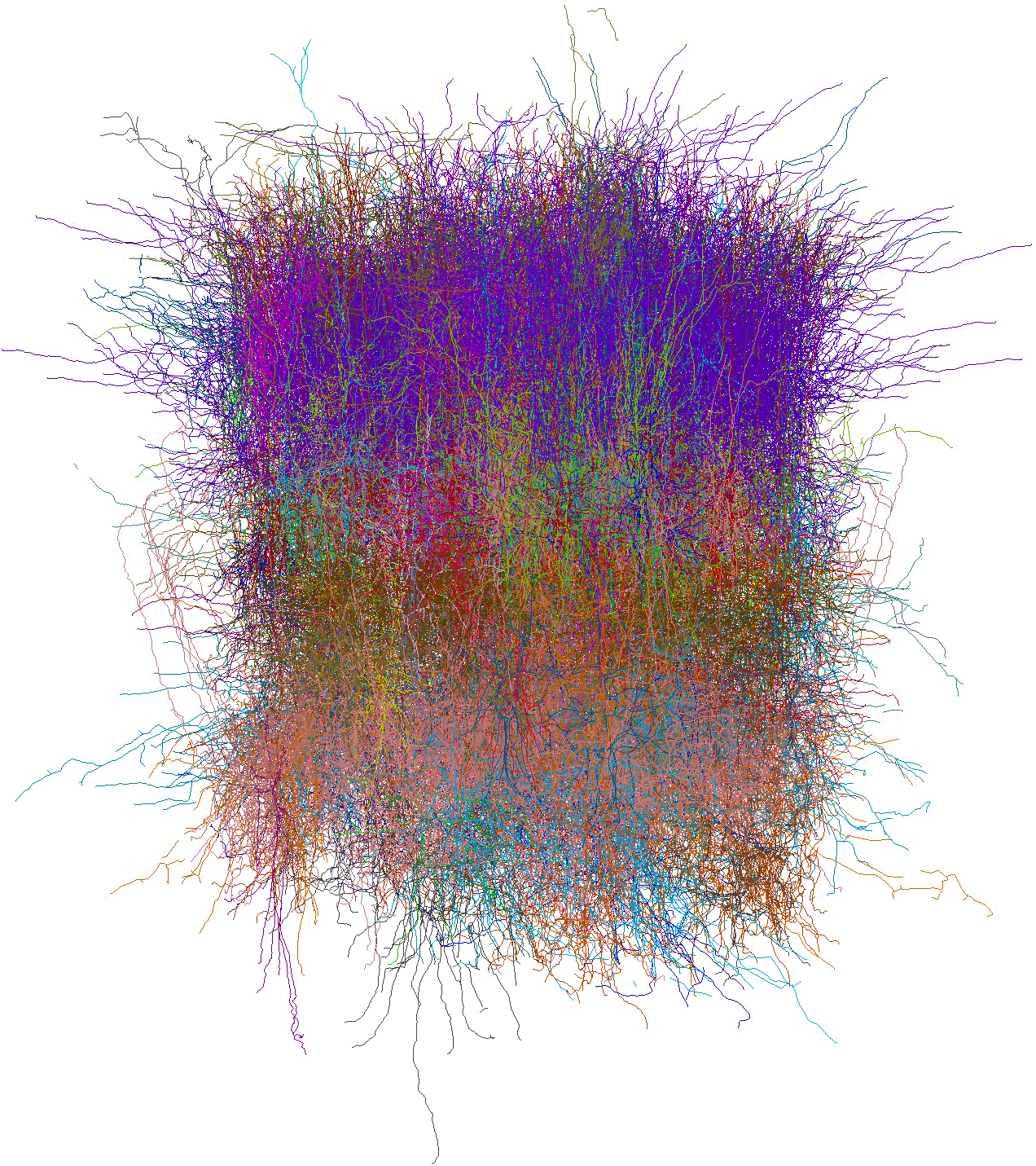
Network structure of the Allen V1 model. Shows biophysical neurons across multiple cortical layers.
Model Overview
Allen V1 model is a biophysical model implemented using BMTK and bionet. This model has the following characteristics:
Morphologically detailed biophysical neurons
Point neurons without morphology (LIF, etc.)
Hierarchical structure with multiple cortical layers
Complex synaptic connection patterns
Model Source
This model is based on an integrated model of mouse primary visual cortex published in the following paper:
Billeh, Y. N., Cai, B., Gratiy, S. L., Dai, K., Iyer, R., Gouwens, N. W., Abbasi-Asl, R., Jia, X., Siegle, J. H., Olsen, S. R., Koch, C., Mihalas, S., & Arkhipov, A. (2020). Systematic Integration of Structural and Functional Data into Multi-scale Models of Mouse Primary Visual Cortex. Neuron, 106(3), 388-403.e18. https://doi.org/10.1016/j.neuron.2020.01.040
This paper presents a network of biophysical models of mouse primary visual cortex that systematically integrates structural and functional data.
bionetlite Coverage
Since bionetlite and Neulite support only biophysical models, we focus on building networks composed of biophysical neurons contained in this model.
Note
The original model contains approximately 52,000 biophysical neurons.
Code Examples
Modifying Existing BMTK Code
To use bionetlite with existing Allen V1 model network construction code, simply change the import statement:
# Before change
# from bmtk.builder.networks import NetworkBuilder
# After change
from bionetlite import NeuliteBuilder as NetworkBuilder
# Following code can be used as-is
net = NetworkBuilder('v1')
# ... network construction code ...
Parallel Execution
Building large-scale networks requires significant computation and time. bionetlite supports parallel execution similar to bionet:
# Parallel execution with 4 processes
mpirun -n 4 python build_network.py
Generated Files
The following files are generated by this model:
V1_population.csv- Information on approximately 52,000 biophysical neuronsV1_V1_connection.csv- Information on approximately 16 million synaptic connectionsProcessed SWC files
Ion channel configuration files
Verifying Results
Note
Please add comparison figures showing execution results from bionetlite+Neulite and bionet+NEURON to this section.
It is recommended to include the following figures:
Raster plot
Firing rate comparison by layer
Execution time comparison
Next Steps
Parallel Execution - Details on parallel execution
Related Links
Allen Institute V1 model: https://brain-map.org/our-research/computational-modeling/models-of-the-mouse-primary-visual-cortex
Allen Cell Types Database: https://celltypes.brain-map.org/
Warning
For complete code and data, please refer to the original Allen Institute model repository.