Neulite Project Documentation
Project Overview
The Neulite project is a system developed to run large-scale biophysical neuron simulations on the supercomputer “Fugaku”. It has achieved simulations of a whole mouse cortex model containing 9 million neurons and 260 billion synapses (see Publication for details).

This document describes Neulite, a high-performance simulation environment for biophysical neuron models, and its frontend bionetlite.
Attention
This documentation is under continuous development. Additional sections and content improvements are planned.
Relationship between BMTK and Neulite
Neulite enables models written in Brain Modeling Toolkit (BMTK) to run with minimal modifications. It leverages existing BMTK models while achieving execution in high-performance computing environments.
Key Features
🚀 Reuse of existing code: Can be used by simply changing the import statements of code written in BMTK
📊 Data standardization: Automatic support for standard formats such as SONATA and Allen Cell Types Database
🔧 Lightweight design: Simple and understandable kernel implementation based on UNIX philosophy
🌐 High portability: Can run on various environments from Raspberry Pi to supercomputers
Components
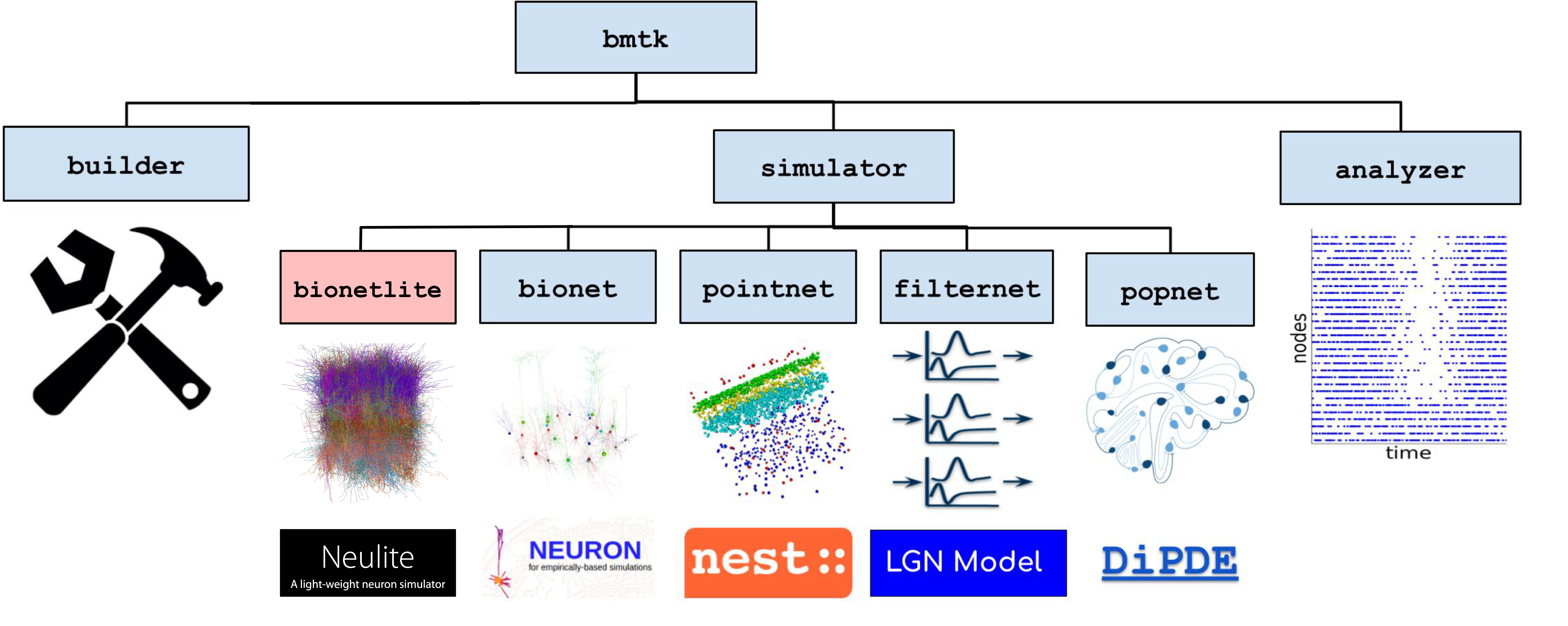
- Bionetlite (Frontend)
A Python package that extends the bionet module of Brain Modeling Toolkit (BMTK). Existing BMTK code can be used with minimal modifications.
- Neulite Kernel (Simulator)
A lightweight and high-performance biophysical neuron simulator. Features a clean codebase implemented in C with a lightweight and extensible design.
Quick Start
# Import bionetlite instead of bionet
from bionetlite import NeuliteBuilder as NetworkBuilder
# Use it the same way as regular bionet
net = NetworkBuilder('v1')
net.add_nodes(...)
net.build()
net.save_nodes(output_dir='network')
net.save_edges(output_dir='network')
Note
For complete setup instructions, please refer to Getting Started.
Documentation Contents
Getting Started
Tutorials
User Guide
Architecture
API Reference
Advanced Topics