Overview
About This Document
This document explains how to use Neulite, a high-performance simulation environment for biophysical neuron models, and its frontend bionetlite.
Neulite enables models written in Brain Modeling Toolkit (BMTK) to run with minimal modifications. It leverages existing BMTK models while achieving execution in high-performance computing environments.
System Architecture
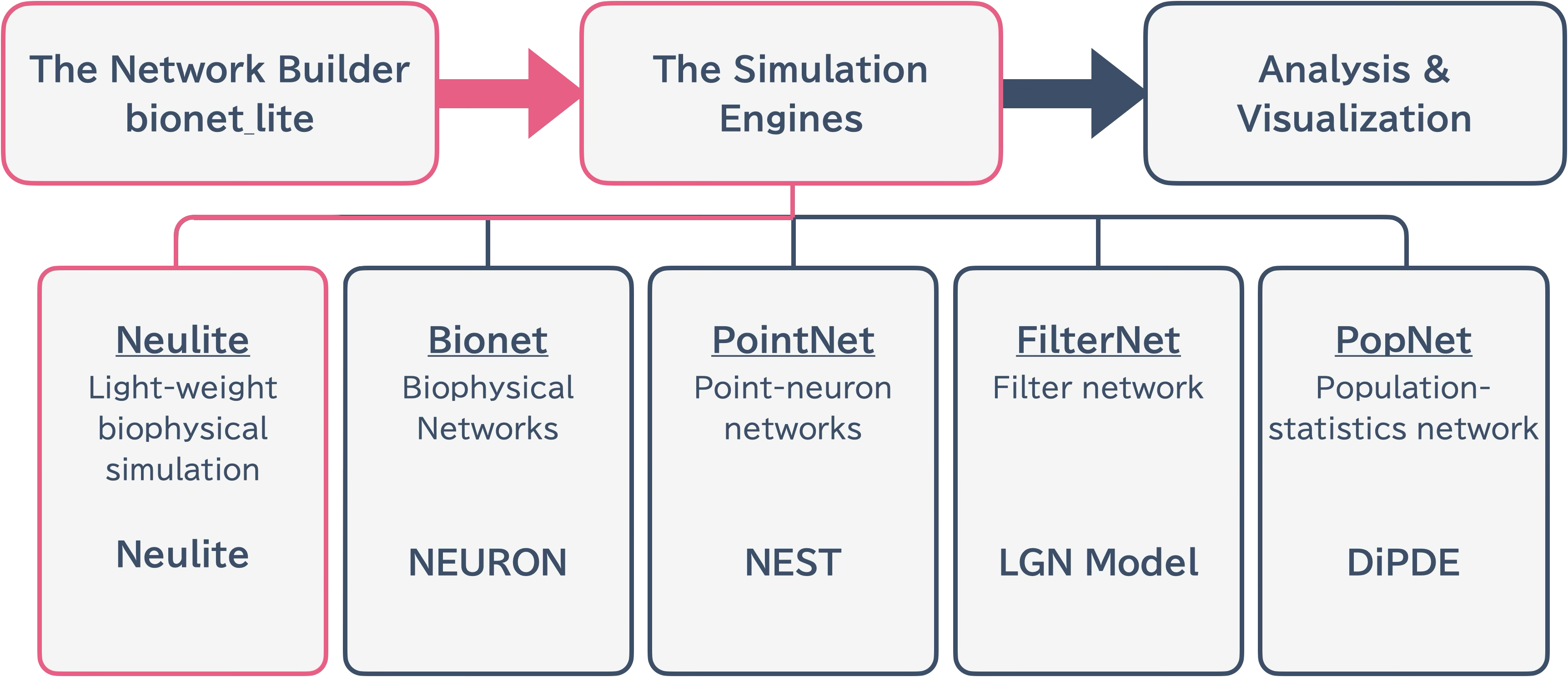
The Neulite system features a clear separation between frontend and backend components. This separation enables flexible network construction in Python while achieving high-speed simulation execution in C.
Backend: Neulite Kernel (C-based)
Frontend: Bionetlite Module (Python-based)
This separation allows network construction and actual simulation execution to be independent, enabling flexible development and maintenance.
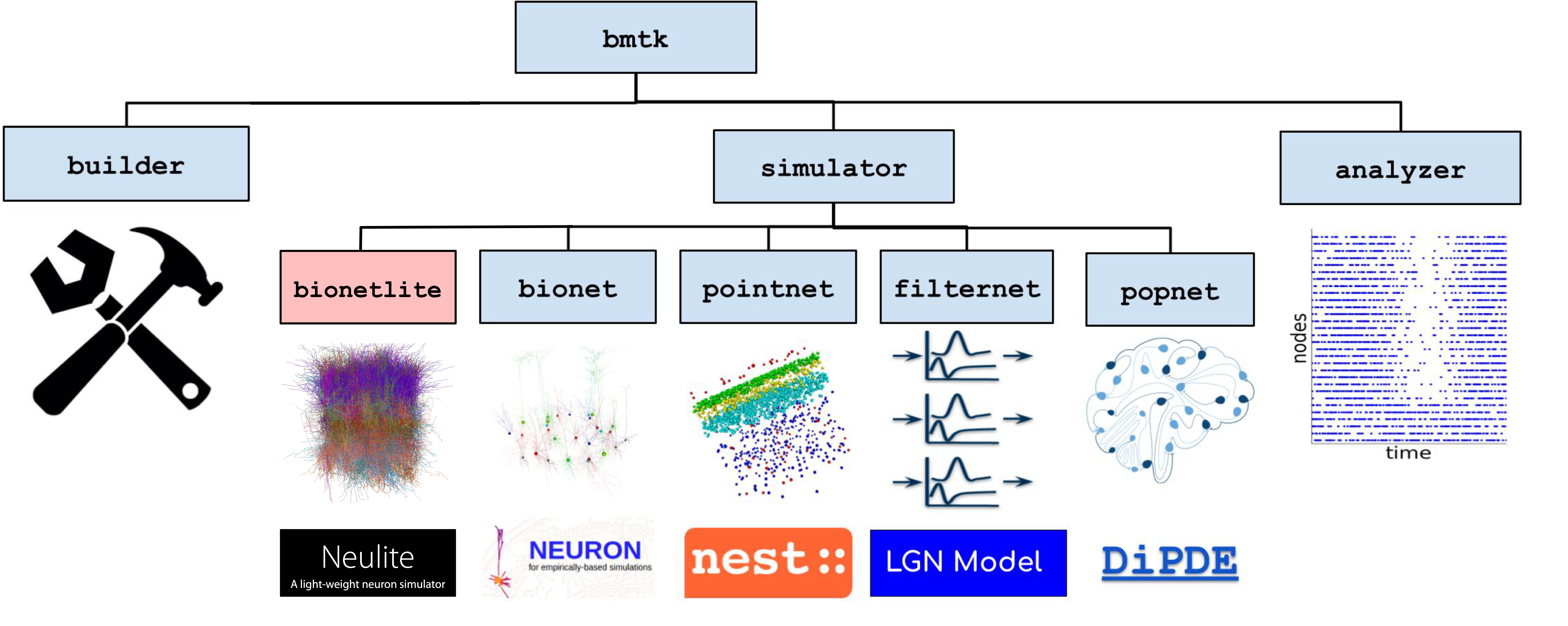
What is Bionetlite?
Bionetlite is a network construction tool compatible with Brain Modeling Toolkit (BMTK). By reusing existing BMTK code with minimal modifications, it automatically supports standard data formats such as SONATA.
Network construction and preprocessing tool implemented in Python
Performs preprocessing of SWC files (axon correction, DFS sorting)
Converts ion channel configuration from JSON to CSV format
Automatically generates configuration files for the Neulite kernel
See What is Bionetlite? for details.
What is Neulite Kernel?
Neulite Kernel is a lightweight simulator for executing large-scale biophysical neuron model simulations at high speed. It specializes in the Perisomatic model from the Allen Cell Types Database and runs across a wide range of environments from Raspberry Pi to the Fugaku supercomputer.
High-speed simulator written in C (C17)
Reads configuration files generated by bionetlite
Users can extend functionality in C
See What is Neulite Kernel? for details.
Usage Flow
Preparation: Prepare BMTK network construction code
Network Construction: Execute Python code that imports bionetlite (configuration files for the Neulite kernel are automatically generated)
Simulation Execution: Run simulation with the Neulite kernel
Result Analysis: Analyze the generated data
Next Steps
What is Neulite Kernel? - Details about Neulite Simulator
What is Bionetlite? - Details and Background of Bionetlite
Getting Started - Setup and Quick Start
Tutorial 1: Steady-State Current Injection in a Single Cell - Practical Tutorials
Overview - Detailed Architecture Explanation